Introduction
Avian orthoavulavirus 1, also known as Newcastle disease virus (NDV), is the etiological agent of a highly contagious viral disease, responsible for significant economic losses in the poultry industry. Many factors should be considered in NDV dissemination, such as lack of farm biosecurity procedures, wild bird population reservoirs, and free movement of migratory birds, people, and animals across borders (Zanetti et al., 2005).
NDV severity varies widely from asymptomatic enteric, lentogenic, and mesogenic to velogenic with neurotropic and viscerotropic variants (Cattoli et al., 2011). Based on genome length and phylogenetic relationships, NDV isolates are classified into two major groups: class I and class II (Miller et al., 2010; Susta, Hamal, Miller, Cardenas-Garcia, et al., 2014). Class I viruses are divided into IX genotypes, which are genetically less diverse with worldwide distribution, are isolated mainly from waterfowl and shorebirds, and are less virulent strains (Dimitrov et al. 2016), except for a single isolate from the Republic of Ireland in 1990 (Diel et al., 2012; Miller et al., 2015).
Class II viruses are both virulent and nonvirulent and infect poultry, pets, and waterfowl birds (Miller et al., 2010; Susta, Hamal, Miller, Cardenas-Garcia, et al., 2014). Class II viruses have been divided into XXI genotypes (Abd Elfatah et al., 2021; Diel et al., 2012; Dimitrov, Abolnik, et al., 2019).
Within class II, genotypes V, VI, VII, and VIII are the most predominant genotypes worldwide (Miller et al., 2009, 2010). Genotype VII viruses are particularly important given the fact that they have been associated with many outbreaks in South America, Asia, Africa, and the Middle East (F. Perozo et al., 2012).
Genotype V can be divided into three sub-genotypes: Va, Vb, and Vc. Sub-genotype Va includes isolates from the United States (US) and Canada, sampled from cormorants and other wild birds such as gulls and pelicans (A. F. Perozo et al., 2008). In Mexico and Central America, the most commonly isolated strains belong to the assigned NDV sub-genotype Vb (Susta, Hamal, Miller, Cardenas-Garcia, et al., 2014).
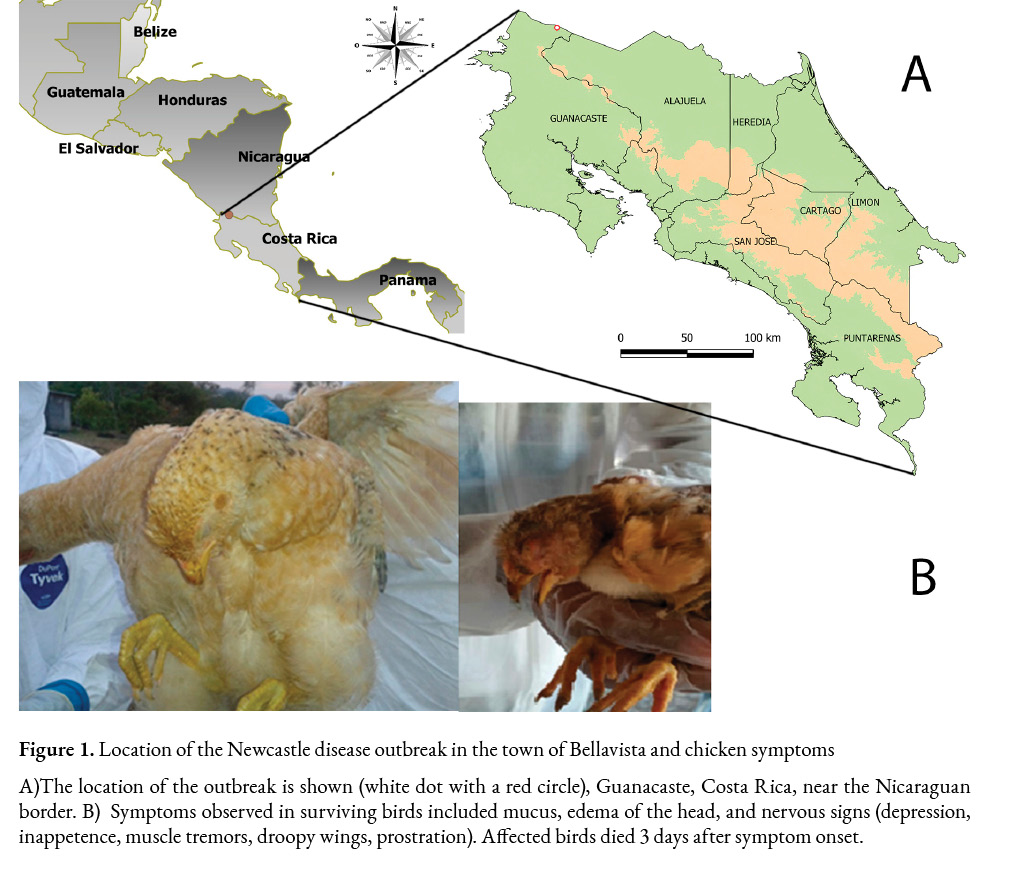
Newcastle disease (ND) is an OIE-notifiable disease and notification of an outbreak in the whole country is mandatory. According to OIE, “A country, zone or compartment may be considered free from Newcastle disease when it has been shown that NDV infection in poultry has not been present in the country, zone or compartment for the last 12 months…” However, if “the infection has occurred in poultry in a free country, zone or compartment, ND free status can be regained three months after a stamping-out policy… is applied, providing that surveillance… has been carried out during that three-month period.” Costa Rica gained its NDV-free status with vaccination according to OIE procedures in 1996 and its declaration as a country free of the velogenic, viscerotropic form of this disease (G/SPS/GEN/119) presented to the World Trade Organization (WTO) in 1999. Non-pathogenic NDV strains had been isolated in Costa Rica since a virulent ND outbreak had been reported in 1990 (Veterinary Services, 2019), and no evidence of virulent Newcastle viruses was circulating in the country until April 22th, 2015, when the regional office of SENASA in Liberia, Guanacaste received a phone call from a backyard chicken owner reporting high morbidity and high mortality in her birds. As part of a passive surveillance and attended by SENASA, the outbreak took place in a remote little rural town named Bellavista, located 83.3 km north of Liberia, near the Nicaraguan border. Sixty-five backyard birds died from a total of 84 unvaccinated exposed animals. The present study aims to report the molecular characterization of a virulent strain of Newcastle disease virus reported in those backyard birds in Costa Rica in 2015.
Materials and methods
Collected samples
The information was collected in situ on April 24th, 2015. The outbreak date was estimated to be February 1st, 80 days before the call was received by SENASA’s regional office in Liberia. The epidemic case was located in Bellavista, Santa Cecilia District, in La Cruz, Guanacaste (11°06’22.0”N 85°20’41.7”W) (Figure 1a). During the visit, the surviving chickens showed respiratory symptoms including cough, swollen head, and watery eyes (Figure 1b), 15 animals were euthanized, and the following samples were taken: 15 serum samples, three pools of 5 samples each of tracheal and cloacal swabs, as well as lungs, trachea, and cecal tonsil tissues. Samples were sent to the National Veterinary Services Laboratory (LANASEVE).
Serological and RT-PCR tests
Serum samples from individuals were analyzed with ID Screen® Influenza A Antibody Competition Multi-species (IDvet, Montpellier, France) ELISA kit, and the Hemagglutinin Inhibition test (HI) was used to detect antibody titers against NDV in sera diluted from 1:8 to 1:1024. HI test was carried out with 4 hemagglutination units (HAU).
For the molecular diagnostics, the samples were extracted with the DNeasy Blood and tissue method (Qiagen, Hilden, Germany Cat number 69506) following the manufacturer’s protocol. A real-time RT-qPCR protocol from the National Veterinary Services Laboratories (NVSL, USDA) was used to detect AIV, targeting the matrix gene (Spackman et al., 2002). Primers used for the matrix gene amplification (influenza virus type A) were: forward M+25 5’-agatgagtcttctaaccgaggtcg-3’, reverse M-124 and 5’-tgcaaaaacatcttcaagtctg-3’ and M-124 SIV, 5’-tgcaaagacactttccagtctctg-3’, at a final concentration of 0.4 μM, M+64 probe 5’-FAM-tcaggccccctcaaagccga-BHQ-1-3’, at a final concentration of 0.12 μM. While a conventional RT-PCR (Stäuber et al., 1995) reverse transcription-polymerase chain reaction (RT-PCR was used for NDV, forward primer 5’-gtcaacatatacacctcatc-3’ and reverse 5’-ggaggatgttggcagcatt-3’, amplifying a 310 bp of the fusion gene region. In both cases, RT-PCRs were adapted to 12.5 µl of reaction using a OneStep RT-PCR Kit (Cat number 210212, Qiagen, Hilden, Germany). NDV RT-PCR concentrations were dNTPs 0.4 µM, RNAse inhibitor 0.04U/µl, buffer OneStep and OneStep Rt-PCR enzyme 1x; both primers were mixed at 0.5 µM, and 2.5 µl of the extracted sample. For Avian Influenza virus (AIV), the reagent concentrations were dNTPs 0.32 µM, RNAse inhibitor 0.04U/µl, buffer OneStep and OneStep RT-PCR enzyme 1X, plus 1.25 mM of MgCl2, primers were at 0.4 µM, and probe at 0.12 µM, and 7.5 µl of the extracted sample; in both RT-PCR the final reaction volume was 12.5 µl. The retrotranscription step cycles were 50 ºC for 30 min, 95 ºC for 15 min, for both viruses followed by 40 cycles at 95 ºC for 15 sec, 57 ºC for 30 sec and 20 sec at 72 ºC and a final extension at 72 ºC for 5 min and kept at 4 °C for NDV. The amplification program for AIV was 40 cycles at 94 ºC for 1 sec and 60 ºC for 31 sec.
Sequencing
All NDV-positive samples were bidirectionally sequenced to confirm the result. PCR products were purified from agarose gel with a QIAquick gel extraction kit (Cat. Number 28704 Qiagen Hilden, Germany) following the manufacturer’s instructions. Sequencing reactions were performed with a BigDye® Terminator cycle sequencing kit v٣.١ (Applied Biosystems, Foster City, CA, USA) using the same primers from the RT-PCR assay at a concentration of ٠.١ µM and 10 µl of purified PCR products, between 15-25 ng/µl in a 20 µl volume reaction. The sequencing products were purified with BigDye XTerminator™ Purification Kit (Applied Biosystems) and subsequently analyzed with an ABI 3130 genetic analyzer (Applied Biosystems).
The following program was used at 96 ° C for 2 min and 30 cycles of 96 ° C for 10 sec, 50 °C for 5 sec, 60 °C for 4 min, and kept at 4 ° C.
Phylogenetic analysis
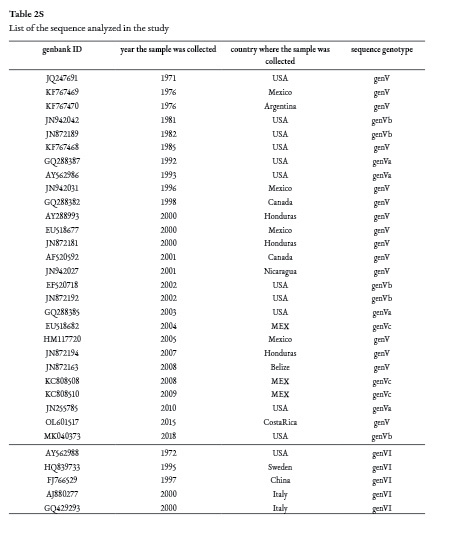
A total of 32 NDV sequences from class II, genotypes V and VI were downloaded from Genbank/ DDBJ /EMBL and were aligned with the Bellavista sequence using Bioedit v7.2 (Hall, 1999). A preliminary tree was generated to determine the temporal signal with IQ-TREE v 1.6.1 (Nguyen et al., 2015) and then analyzed with TempEst (Rambaut et al., 2016). A BEAUti XML file was designed with the following parameters, the general time reversible model with empirical base frequencies (GTR+F) considered the BEAST substitution model according to IQ-TREE; the uncorrelated relaxed clock model was chosen (using marginal likelihood estimation (MLE) path sampling (PS) with 100 steps and 1 million chains and Bayes factor approaches) (Baele et al., 2013); and finally the coalescent constant size model tree was selected. A tree was generated with 30 million iterations using the Monte Carlo Markov chains (MCMC) method in BEAST1.10 (Suchard et al., 2018).
The Maximum Clade Credibility (MCC) tree was created using TreeAnnotator (Bouckaert et al., 2019).
Results
Serology tests
No antibodies were detected against the AIV virus using the ID Screen® ELISA kit. The haemagglutination inhibition test (HI) showed a titer range of antibodies against NDV from negative to 1024. From the 15 serum samples received, 13 had titers above 1:16 HAU, which are considered positive, ranging from 1:256 to 1:1024.
Molecular tests
All samples were negative for the Avian influenza virus (AIV). However, lungs, tracheae tissue, and cloacal and tracheal swab samples and their 1:10 dilutions were positive for NDV (Figure 2a).
The NDV sequences obtained were identical and the 310 bp sequence was deposited in the Genbank database under the accession number OL601517. The amino acid sequence of the protease cleavage site within the amplicon matched a virulent strain pattern (112RRQKRF117) (Figure 2b).
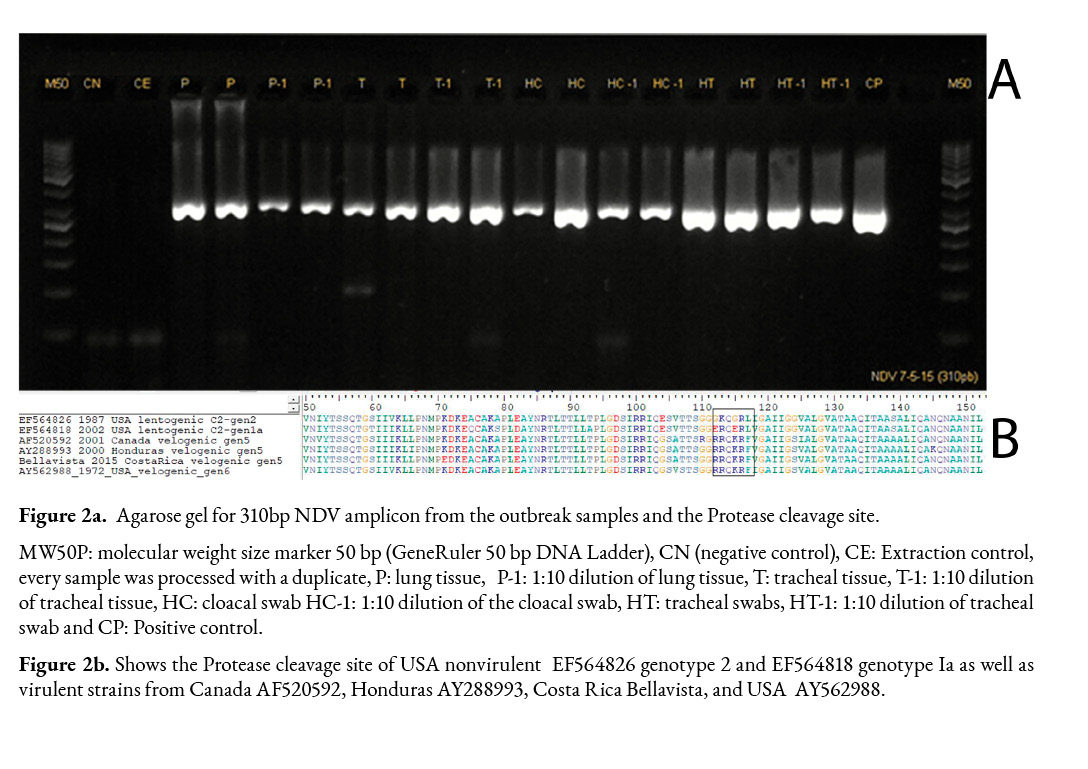
According to BLASTN (Agarwala et al., 2018) the Bellavista nucleotide sequence had a 98.7% identity and an e value of 2e-152 with genotype V velogenic sequences from Belize (KF767467, JN942045, KF767466, MK214318, MH392217) and Honduras (JN872194), which were collected in 2008 and 2007, respectively.
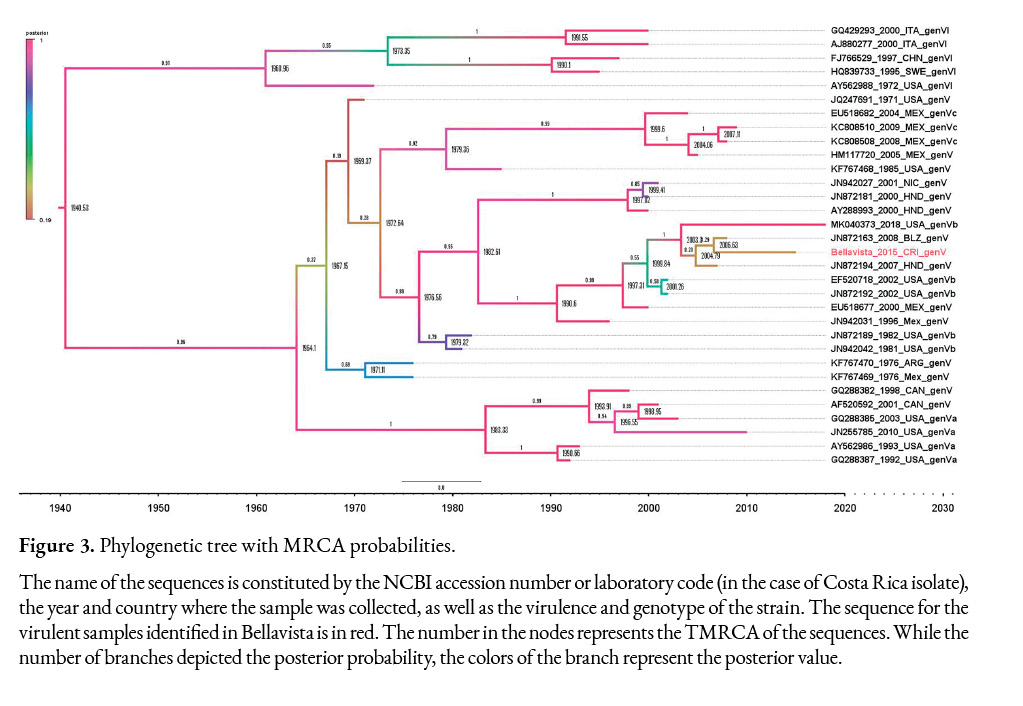
Figure 3 shows the tree built with the BEAST software. The Effective Sample Size (ESS) for all parameters was greater than 751, which indicates that the model has converged.
Phylogenetic analysis
The phylogenetic analysis depicted the relationship among the selected sequences as well as the time of the most recent common ancestor (TMRCA) in the nodes. The tree from Figure 3 shows the posterior value in the branches. According to the sequences available from Central America, Mexico, USA, Canada, and Argentina, which were deposited in Genbank/ DDBJ /EMBL, the common ancestor that originated the Bellavista-2015 (OL601517), Belize-2008, Honduras-2007, and USA-2018 sequences (98.71% to 96-77% identity with Costa Rican sequence), dated from 2003 high posterior density (HPD) interval 95% (1955-2006). These sequences share a common ancestor with two sequences from the USA collected in 2002, and with the sequence EU518677 from Mexico-2000, which has a 98.06% identity with the virulent Costa Rican sequence; the TMRCA among these sequences was 1990 (95% HPD 1984-1994) with a 0.87 probability that this ancestor came from the USA. All these sequences are classified as genotype Vb.
If the sequences from 2000-2001 isolated in Honduras and Nicaragua, respectively, are included with the rest of the Central American sequences, the TMRCA of these sequences is 1982, 95% HPD (1975-1990), and their probability of coming from the USA is 0.88, the probability that the sequences belonging to genotype Vb came from the USA is indeed 1.0. Considering these results and the sequences analyzed here, NDV ancestors of genotype Vb related to Central American and Mexican outbreaks could have been circulating in the USA since 1976 with 95% HPD (1971-1980) and then spread to these countries.
Discussion
Given the susceptibility of backyard birds to acquire viral infections by the exotic Newcastle disease virus, the lack of vaccines against this disease and of biosecurity measures, birds smuggling, and the greater exposure of backyard birds to wild birds, SENASA maintains active and passive surveillance in backyard poultry farms throughout the year in the areas of greater epidemiological risk. SENASA has selected geographic areas that are considered to be at greater risk due to their proximity to national parks, biological reserves, coastal areas, borders, areas of high poultry density, as well as points of entry for people and goods into the national territory, such as ports and airports. In addition, through newsletters and informational brochures, SENASA constantly motivates veterinarians and bird owners in general to report any symptoms compatible with exotic NDV according to Executive Decree No. 34669-MAG, List of notifiable animal diseases. Once a notification of a suspected case is received, it is investigated and attended by the SENASA staff responsible for the affected area, documenting the collection of diagnostic samples, as well as the sanitary measures implemented while waiting for the results of the National Veterinary Services Laboratory (LANASEVE). As a result of the NDV surveillance program in commercial and backyard birds in Costa Rica, Lentogenic strains of NDV have been isolated in SPF embryonic eggs and sequenced since 2007 (León-Rodríguez et al., 2009), detecting only the vaccine strains La Sota and B1 (genotype II), and PHY-LMV42 (genotype Ia). It was not until 2015 when a virulent strain was diagnozed in backyard chickens near the Nicaraguan border.
The symptoms shown by the infected birds in the town of Bellavista, the lethality rate calculated at 77%, and the 95% morbidity were not compatible with ND or highly pathogenic avian influenza (HPAI) or even other etiologies; for this reason, a rapid and accurate diagnosis was crucial.
Serological tests (first results obtained) showed high titers of antibodies against NDV in most of the analyzed samples, which is relevant since the birds were not vaccinated. No presence of antibodies to AIV was detected. In Costa Rica, no antibodies against the influenza virus were detected in 151 backyard chickens from 13 flocks during three separate periods in July 2005, November 2005, and February 2007 (Hernandez-Divers et al., 2008). Unfortunately, no other studies have been done in the country on poultry, either backyard or wild birds.
The positive ND serological result was supported by molecular tests, which confirmed the absence of AIV by real-time RT-PCR and the presence of NDV by conventional RT-PCR. The NDVconventional RT-PCR assay can detect between 5 x 102 EID50 in live vaccine preparations and 105 EID50 or 0.056 haemagglutinating units of NDV in the inactivated vaccine (Stäuber et al., 1995). These results were confirmed by the sequencing process as a virulent strain belonging to genotype Vb.
From 2005 to 2021, there have been 30 ND outbreaks in the area, 9 in Mexico and 21 in Central America, including the one from Costa Rica in 2015 and 5 in Nicaragua, which were reported in 2011, 2012, 2013, 2015, and 2016 (Table 1S).
Based on the phylogenetic analysis, the velogenic NDV strain isolated in Costa Rica belongs to genotype Vb. Genotype V caused outbreaks in Europe and the USA in 1970 (Ballagi-Pordány et al., 1996). Considering the sequences evaluated here, the TMRCA of genotype Vb could have been circulating since 1976 with 95% HPD (1971-1980) in the Americas. The NDV genotype Vb outbreak caused in the USA during 2002-2003 was closely related to cases in Honduras in 2000 and Mexico in 1996 (Dimitrov, Ferreira, et al., 2019; Susta, Hamal, Miller, Cardenas-Garcia, et al., 2014).
Based on our results, the virulent sequences obtained in the Central American countries could have come from Mexico. The first virulent NDV reported in Mexico was in 1946, which was detected in 1-day-old chicks imported from the United States of America (Garcia et al., 2013; Merino et al., 2009). In January 2009, NDV was identified in Tecpatán and Cintalapa, Chiapas, with lethality rates ranging from 40% to 80% (Garcia et al., 2013), lower than described in the Bellavista outbreak.
After eradicating NDV in Costa Rica, every backyard bird in town and its surroundings (1 km) was euthanized, for a total of 3604 birds, including 3495 chickens, 66 turkeys, 6 geese, and 37 ducks. In April 2017, Costa Rica recovered its disease-free status through the promulgation of an executive decree (Decreto 40301-MAG, 2017).
Belize reported the presence of the virus every year from 2015 to 2021, Mexico in 2016 and 2019, Nicaragua in 2016, and Honduras in 2019. Considering this situation, the reintroduction of virulent NDV in Costa Rica is quite possible; therefore, continuous surveillance of morbidity and mortality of commercial backyard poultry is fundamental. During this surveillance over the last six years, only NDV lentogenic strains have been detected.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgments
The authors thank Dr. Ricardo Salazar-Aragón who attended the case and took the samples and all the SENASA personnel (administrative and field staff) who worked in the eradication of this outbreak.
References
Abd Elfatah, K. S., Elabasy, M. A., El-khyate, F., Elmahallawy, E. K., Mosad, S. M., El-Gohary, F. A., Abdo, W., Al-Brakati, A., Seadawy, M. G., Tahoon, A. E., & El-Gohary, A. E. (2021). Molecular Characterization of Velogenic Newcastle Disease Virus (Sub-Genotype VII.1.1) from Wild Birds, with Assessment of Its Pathogenicity in Susceptible Chickens. Animals, 11(2), 505. https://doi.org/10.3390/ani11020505
Agarwala, R., Barrett, T., Beck, J., Benson, D. A., Bollin, C., Bolton, E., Bourexis, D., Brister, J. R., Bryant, S. H., Canese, K., Cavanaugh, M., Charowhas, C., Clark, K., Dondoshansky, I., Feolo, M., Fitzpatrick, L., Funk, K., Geer, L. Y., Gorelenkov, V., … Zbicz, K. (2018). Database resources of the National Center for Biotechnology Information. Nucleic Acids Research, 46(D1), D8–D13. https://doi.org/10.1093/nar/gkx1095
Decreto 40301-MAG, (2017).
Baele, G., Lemey, P., & Vansteelandt, S. (2013). Make the most of your samples: Bayes factor estimators for high-dimensional models of sequence evolution. BMC Bioinformatics, 14(1), 85. https://doi.org/10.1186/1471-2105-14-85
Ballagi-Pordány, a, Wehmann, E., Herczeg, J., Belák, S., & Lomniczi, B. (1996). Identification and grouping of Newcastle disease virus strains by restriction site analysis of a region from the F gene. Archives of Virology, 141(February), 243–261. https://doi.org/10.1007/BF01718397
Bouckaert, R., Vaughan, T. G., Barido-Sottani, J., Duchêne, S., Fourment, M., Gavryushkina, A., Heled, J., Jones, G., Kühnert, D., De Maio, N., Matschiner, M., Mendes, F. K., Müller, N. F., Ogilvie, H. A., du Plessis, L., Popinga, A., Rambaut, A., Rasmussen, D., Siveroni, I., … Drummond, A. J. (2019). BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLOS Computational Biology, 15(4), e1006650. https://doi.org/10.1371/journal.pcbi.1006650
Cattoli, G., Susta, L., Terregino, C., & Brown, C. (2011). Newcastle disease a review of field recognition and current methods of laboratory detection. Journal of Veterinary Diagnostic Investigation, 23(4), 637–656. https://doi.org/10.1177/1040638711407887
Diel, D. G., da Silva, L. H. A., Liu, H., Wang, Z., Miller, P. J., & Afonso, C. L. (2012). Genetic diversity of avian paramyxovirus type 1: Proposal for a unified nomenclature and classification system of Newcastle disease virus genotypes. Infection, Genetics and Evolution, 12(8), 1770–1779. https://doi.org/10.1016/j.meegid.2012.07.012
Dimitrov, K. M., Abolnik, C., Afonso, C. L., Albina, E., Bahl, J., Berg, M., Briand, F. X., Brown, I. H., Choi, K. S., Chvala, I., Diel, D. G., Durr, P. A., Ferreira, H. L., Fusaro, A., Gil, P., Goujgoulova, G. V., Grund, C., Hicks, J. T., Joannis, T. M., … Wong, F. Y. K. (2019). Updated unified phylogenetic classification system and revised nomenclature for Newcastle disease virus. Infection, Genetics and Evolution, 74(June). https://doi.org/10.1016/j.meegid.2019.103917
Dimitrov, K. M., Ferreira, H. L., Pantin-Jackwood, M. J., Taylor, T. L., Goraichuk, I. V., Crossley, B. M., Killian, M. L., Bergeson, N. H., Torchetti, M. K., Afonso, C. L., & Suarez, D. L. (2019). Pathogenicity and transmission of virulent Newcastle disease virus from the 2018–2019 California outbreak and related viruses in young and adult chickens. Virology, 531(January), 203–218. https://doi.org/10.1016/j.virol.2019.03.010
Garcia, S. C., Navarro Lopez, R., Morales, R., Olvera, M. A., Marquez, M. A., Merino, R., Miller, P. J., & Afonsoa, C. L. (2013). Molecular epidemiology of newcastle disease in Mexico and the potential spillover of viruses from poultry into wild bird species. Applied and Environmental Microbiology, 79(16), 4985–4992. https://doi.org/10.1128/AEM.00993-13
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp., 41, 95–98. http://jwbrown.mbio.ncsu.edu/JWB/papers/1999Hall1.pdf
Hernandez-Divers, S. M., Villegas, P., Jimenez, C., Hernandez-Divers, S. J., Garcia, M., Riblet, S. M., Carroll, C. R., O’Connor, B. M., Webb, J. L., Yabsley, M. J., Williams, S. M., & Sanchez, S. (2008). Backyard chicken flocks pose a disease risk for neotropic birds in Costa Rica. Avian Diseases, 52(4), 558–566. https://doi.org/10.1637/8298-032808-Reg.1
León-Rodríguez, B., Vargas-Brenes, O., Guevara Soto, M., & Solano-Pereira, M. (2009). Análisis molecular de una cepa de virus de Newcastle de origen vacunal aislada a partir de un hisopado cloacal de aves sanas en Costa Rica - Molecular analysis of an isolated Newcastle disease virus strain obtained from cloacal swabs in healthy poultry fa. REDVET. Revista Electrónica de Veterinaria, 10(11), 1–19.
Merino, R., Villegas, H., Quintana, J. A., & Calderon, N. (2009). Characterization of Newcastle disease viruses isolated from chicken, gamefowl, pigeon and quail in Mexico. Veterinary Research Communications, 33(8), 1023–1030. https://doi.org/10.1007/s11259-009-9321-5
Miller, P. J., Decanini, E. L., & Afonso, C. L. (2010). Newcastle disease: Evolution of genotypes and the related diagnostic challenges. Infection, Genetics and Evolution, 10(1), 26–35. https://doi.org/10.1016/j.meegid.2009.09.012
Miller, P. J., Haddas, R., Simanov, L., Lublin, A., Rehmani, S. F., Wajid, A., Bibi, T., Khan, T. A., Yaqub, T., Setiyaningsih, S., & Afonso, C. L. (2015). Identification of new sub-genotypes of virulent Newcastle disease virus with potential panzootic features. Infection, Genetics and Evolution, 29(July), 216–229. https://doi.org/10.1016/j.meegid.2014.10.032
Miller, P. J., Kim, L. M., Ip, H. S., & Afonso, C. L. (2009). Evolutionary dynamics of Newcastle disease virus. Virology, 391(1), 64–72. https://doi.org/10.1016/j.virol.2009.05.033
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., & Minh, B. Q. (2015). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution, 32(1), 268–274. https://doi.org/10.1093/molbev/msu300
Perozo, A. F., Merino, R., Afonso, C. L., Villegas, P., Calderon, N., Perozo, F., Merino, A. R., Afonso, B. C. L., Villegas, C. P., & Calderon, N. B. (2008). Biological and Phylogenetic Characterization of Virulent Newcastle Disease Virus Circulating in Mexico Biological and Phylogenetic Characterization of Virulent Newcastle Disease Virus Circulating in Mexico. Avian Diseases, 52(3), 472–479. https://doi.org/10.1637/1933-5334(2008)3
Perozo, F., Marcano, R., & Afonso, C. L. (2012). Biological and phylogenetic characterization of a genotype VII Newcastle disease virus from Venezuela: Efficacy of field vaccination. Journal of Clinical Microbiology, 50(4), 1204–1208. https://doi.org/10.1128/JCM.06506-11
Rambaut, A., Lam, T. T., Max Carvalho, L., & Pybus, O. G. (2016). Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evolution, 2(1), vew007. https://doi.org/10.1093/ve/vew007
Spackman, E., Senne, D. A., Myers, T. J., Bulaga, L. L., Garber, L. P., Perdue, M. L., Lohman, K., Daum, L. T., & Suarez, D. L. (2002). Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. Journal of Clinical Microbiology, 40(9), 3256–3260. https://doi.org/10.1128/JCM.40.9.3256
Stäuber, N., Brechtbühl, K., Bruckner, L., & Hofmann, M. A. (1995). Detection of Newcastle disease virus in poultry vaccines using the polymerase chain reaction and direct sequencing of amplified cDNA. Vaccine, 13(4), 360–364. http://www.ncbi.nlm.nih.gov/pubmed/7793131
Suchard, M. A., Lemey, P., Baele, G., Ayres, D. L., Drummond, A. J., & Rambaut, A. (2018). Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evolution, 4(1), 1–5. https://doi.org/10.1093/ve/vey016
Susta, L., Hamal, K. R., Miller, P. J., Cardenas-Garcia, S., Brown, C. C., Pedersen, J. C., Gongora, V., & Afonso, C. L. (2014). Separate evolution of virulent Newcastle disease viruses from Mexico and Central America. Journal of Clinical Microbiology, 52(5), 1382–1390. https://doi.org/10.1128/JCM.00066-14
Susta, L., Hamal, K. R., Miller, P. J., Cardenas-garcia, S., Brown, C. C., Pedersen, J. C., Gongora, V., & Afonso, L. (2014). Separate Evolution of Virulent Newcastle Disease Viruses from Mexico and Central America. J Clin Microbiol, 52(5), 1382–1390. https://doi.org/10.1128/JCM.00066-14
Veterinary Services, A. and P. H. I. (2019). Report on the Review of Costa Rica’s Animal Health Statuses (Issue August).
Zanetti, F., Berinstein, A., Pereda, A., Taboga, O., Carrillo, E., Zanetti, F., Berinstein, A. A., Pereda, A. B. A., Taboga, A. B. O., & Carrilloabc, E. (2005). Molecular Characterization and Phylogenetic Analysis of Newcastle Disease Virus Isolates from Healthy Wild Birds. Avian Diseases, 49(4), 546–550.

Licencia Creative Commons Atribución-No-Comercial
SinDerivadas 3.0 Costa Rica
Supplementary material
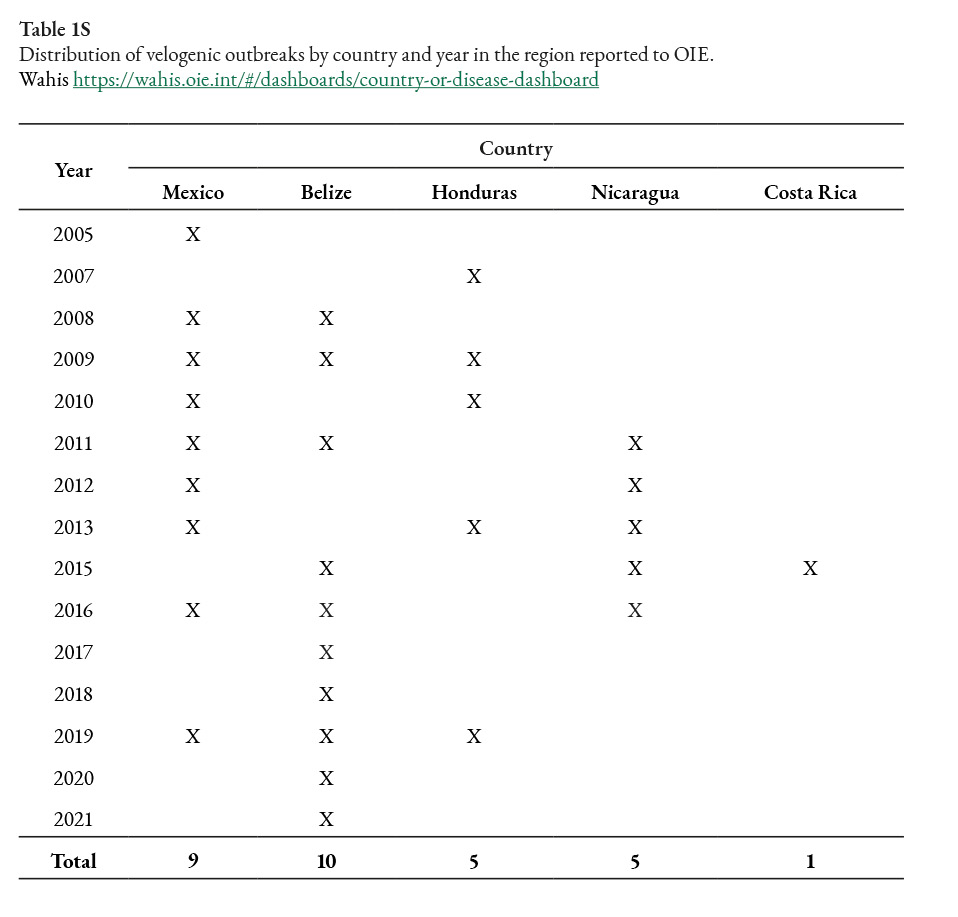
MLE
Path sampling log marginal likelihood
Strict clock = -1454.9649814577474
Uncorrelated relaxed clock =-1365,93992878

 , Juan M. Cordero-Solórzano2, Idania Chacón1, Olga Aguilar1, Guisella Chaves1, Mónica Guzmán1, Fabián Carvajal1, Ronaldo Chaves3
, Juan M. Cordero-Solórzano2, Idania Chacón1, Olga Aguilar1, Guisella Chaves1, Mónica Guzmán1, Fabián Carvajal1, Ronaldo Chaves3 0000-0002-0070-6746, idania.chacon.g@senasa.go.cr
0000-0002-0070-6746, idania.chacon.g@senasa.go.cr  0000-0003-4790-0957, olga.aguilar.a@senasa.go.cr
0000-0003-4790-0957, olga.aguilar.a@senasa.go.cr  0000-0001-5284-0739, guisella.chaves.g@senasa.go.cr
0000-0001-5284-0739, guisella.chaves.g@senasa.go.cr  0000-0002-6911-8715, monica.guzman.s@senasa.go.cr,
0000-0002-6911-8715, monica.guzman.s@senasa.go.cr,  0000-0002-0363-7248, fabian.carvajal.d@senasa.go.cr
0000-0002-0363-7248, fabian.carvajal.d@senasa.go.cr  0000-0001-6030-7686
0000-0001-6030-7686 0000-0002-0641-4998
0000-0002-0641-4998 0000-0001-9703-7747
0000-0001-9703-7747 Autor de correspondencia: bernal.leon.r@senasa.go.cr
Autor de correspondencia: bernal.leon.r@senasa.go.cr




